|
Content
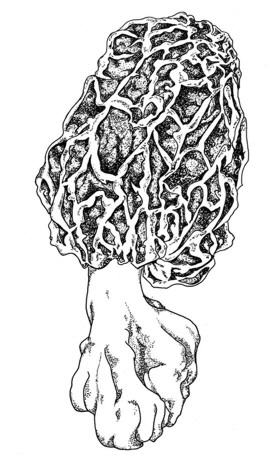
1. What are morels?
2. How many species?
3. Habitats
4. Research aims
5. Methods
6. Results
7. Suggested readings
Morchella spongiola (Boud.),
Journalcover of Organisms Diversity & Evolution 5, 2005 by Patricia Luis
1. What are morels? - an introduction
Morels are widely known as eatable, early appearing and therefore highly priced fungi, were several attempts were undertaken to cultivate them. Morels belong to the Ascomycota, order: Pezizales, family: Morchellaceae. One major characteristic of this family is the coenosity of the spores with sometimes more than 20 nuclei (Fig. 1). Further informations are available at Wikipedia [english] [german].
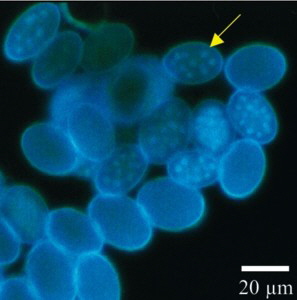 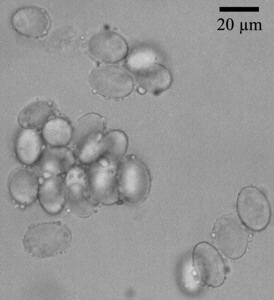
Fig. 1. Morchella spores stained using DAPI, thus making the nuclei visible and without staining.
2. How many species?
The debate about species concepts and diversity in the genus Morchella Dill. ex Pers. is very old and longstanding. In systematic treatments essentially based on morphological features, the species number in Morchella fluctuates between 3 and 30, reflecting the enormous typological variability of the fruit bodies according to habitats and their specific abiotic and biotic conditions. Through upcoming molecular methods, i.e. amplifying and analysing of genomic DNA, several systematic questions could be answered. Sequence data of nuclear ribosomal DNA confirmed the monophyly of the Morchellaceae and the separation of Adnatae (yellow morels) and Distantes (black morels) made by Boudier (1897) in one of the first systematic treatments of the genus. The huge variability of macroscopic features (see Fig. 2) and the consistence of microscopic ones found for spores and hyphae hamper the clear distinction of more than three species in field surveys. This is reflected by the fact that most recent works only distinguish three species within Germany: Morchella elata Fr. (including M. conica Pers., M. costata (Vent.) Pers., M. deliciosa Fr., M. hortensis Boud.), Morchella esculenta (L.) Pers. (including M. crassipes (Vent.) Pers. : Fr., M. rotunda (Fr.) Boud., M. umbrina Boud., M. spongiola Boud., M. vulgaris (Pers.) Boud.) and Morchella gigas (Batsch : Fr.) Pers. (including M. hybrida Pers., M. rimosipes DC., Mitrophora semilibera (DC. : Fr.) Lev.).

Fig. 2. Morchella spongiola collected at the same site (Jena, Kunitz, 22.4.2004) showing a high
variability of the fruitbody through different life stages and site conditions. The ITS sequences
of these morels were 100% identical (see also Masaphy et al. 2010 for more informations on this topic).
Current work and progress in systematics, biology, ecology and biogeography you will find in the Morel Data Collection Project (MDCP), provided from Michael Kuo, Kerry O´Donnell and Carol Carter and many other people here [Link].
3. Habitats
Morels are growing in a broad range of different habitats. The habitats seems to differ in their ecological stability. Thus, morels grow on one hand in massively disturbed or antropogenic habitats like burned forest sites or mulched gardens and on other hand in “relatively stable” ecosystems like sand dunes (near to Pinus forests), orchards, pioneer- or floodplain forests.
 
Fig. 3. Morchella spongiola collected at Reinsdorf in an orchard (2006).
4. Research aims
First aim was to clarify the phylogeny of the Morchellaceae. We wanted to know how many genetic lineages could be observed in Central Europe before we could answer ecological questions. Thereafter we investigated the genetic and functional diversity of laccase genes in order to proof potential soil organic matter turnover, i.e. litter degradation processes of several species.
5. Methods
In order to investigate the phylogeny or biogeography of different morels and further members of the Morchellaceae family, standard molecular methods were used. In particular, DNA was extracted from collected fruitbodies or monospore cultures and thereafter used for PCR amplification of genes coding for ribosomal RNA or spacer regions, i.e. the internal transcribed spacer (ITS) (Fig. 4, Table 1). The amplified gene fragments were cloned and subsequently sequenced. Afterwards the sequences were aligned and used in phylogenetic programs to analyse their taxonomic relationships.
Moreover laccase-like multicopper oxidases and their activity were analysed according the methods refered under the Laccase Project section.
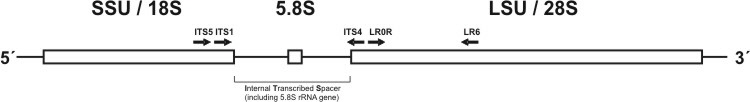
Fig. 4. Scheme of one tandem repeat encoding ribosomal genes and including internal transcribed spacer region (ITS). Primers used in this study are shown.
Table 1. Primer paires used in this study, according White et al. 1990, Moncalvo et al. 1995, Vilgalys&Hester 1990
|
Primer name
|
Direction
|
Sequence 5´-3´
|
|
ITS1
|
forward
|
TCCGTAGGTGAACCTGCGG
|
|
ITS4
|
reverse
|
TCCTCCGCTTATTGATATGC
|
|
ITS5
|
forward
|
GGAAGTAAAAGTCGTAACAAGG
|
|
LR0R
|
forward
|
ACCCGCTGAACTTAAGC
|
|
LR6
|
reverse
|
CGCCAGTTCTGCTTACC
|
|
6. Results
Several results were obtained and provided at separate pages (result section I and II).
Section I: Phylogeny using ITS and LSU sequences [Link]
Section II: Biogeography of yellow morels and analysis of laccase genes [Link]
7. Suggested readings
Kellner H., Luis P., Buscot F. (2007): Diversity of laccase-like multicopper oxidase (LMCO) genes in Morchellaceae: identification of genes potentially involved in extracellular activities related to plant litter decay. FEMS Microbiology Ecology 61: 153-163.
Kellner H., Renker C., Buscot F. (2005): Species diversity within Morchella esculenta group (Ascomycota: Morchellaceae) in Germany and France. Organisms Diversity & Evolution 5: 101-107.
Kuo M. (2006): Morel Data Collection Project. Retrieved from the MushroomExpert.Com Web site: http://www.mushroomexpert.com/mdcp/index.html
Masaphy S., Zabari L., Goldberg D., Jander-Shagug. (2010): The Complexity of Morchella Systematics: A Case of the Yellow Morel from Israel. Fungi 3:14-18.
Moncalvo J.M., Wang H.-H., Hseu R.-S. (1995): Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia 87: 223-238.
O’Donnell K., Cigelnik E., Weber S.N., Trappe J.M. (1997): Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia 89: 48-65.
O'Donnell K, Rooney AP, Mills GL, Kuo M, Weber NS, Rehner SA (2011): Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genetics and Biology 48: 252–265.
Vilgalys R., Hester M. (1990): Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172: 4238-4246.
White T.J., Bruns T., Lee S., Taylor J. (1990): Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. (Eds.): PCR protocols: a Guide to Methods and Applications. Academic Press, San Diego, pp. 315–322.
Wipf D., Fribourg A., Munch J.-C., Botton B., Buscot F. (1999): Diversity of the internal transcribed spacer of DNA in morels. Can J Microbiol 45: 769-778.
Acknowledgement
I would like to thank Dr. C. Renker and Dr. P. Luis for their help during this study. Furthermore Bettina Schlitt for help in laboratory work. Many thanks also to many people, who helped in providing vouchers or for their helpful commentaries.
|