1. What are fungal laccases?
Laccases (benzenediol: oxygen oxidoreductase, EC 1.10.3.2) catalyze the oxidation of various aromatic, particularly phenolic substrates (e.g. hydroquinone, guaiacol, 2,6-dimethoxyphenol or phenylene diamine), coupled to the reduction of molecular oxygen to water. Laccases as well as ascorbate oxidases (EC 1.10.3.3) and ceruloplasmins/ferroxidase (EC 1.16.3.1) usually contain several copper atoms in the catalytic centre. Therefore, they belong to the enzyme superfamily of multicopper oxidases, which is a widely distributed protein family among pro- and eukaryotes. However, laccases or laccase-like multicopper oxidases (LMCO) were predominantly described in fungi and plants, were they occur as multigene family with sometimes more than 10 different laccase genes. For example, the basidiomycete Coprinopsis cinerea and the plant Arabidopsis thaliana have both 17 different genes (see also Hoegger et al. 2006 for more details).
 Due to their large capacities to oxidize polyphenols, fungal laccases are involved in several biochemical processes. In the environment they are involved in the biodegradation of lignin and humic acids (depolymerization) and biosynthesis of new humic compounds (polymerization). Thus laccases of litter and wood degrading fungi strongly contribute to soil organic matter turnover and the global carbon cycle. Fungal laccases also play an important role in physiological processes related to pathogenesis, morphogenesis, i.e. fruitbody development, pigmentation and to cell detoxification. While laccases involved in lignin degradation are extracellular enzymes, the ones responsible for the other processes essentially act intracellularly. Due to the oxidation of a variety of substrates, fungal laccases especially from basidiomycetes receive also attention for use in diverse biotechnological applications, like bioremediation. Laccases are involved in the biotransformation of many persistent environmental pollutants, like azo dyes, polycyclic aromatic hydrocarbons, endocrine disruptors or polycyclic musk fragrances.
Due to their large capacities to oxidize polyphenols, fungal laccases are involved in several biochemical processes. In the environment they are involved in the biodegradation of lignin and humic acids (depolymerization) and biosynthesis of new humic compounds (polymerization). Thus laccases of litter and wood degrading fungi strongly contribute to soil organic matter turnover and the global carbon cycle. Fungal laccases also play an important role in physiological processes related to pathogenesis, morphogenesis, i.e. fruitbody development, pigmentation and to cell detoxification. While laccases involved in lignin degradation are extracellular enzymes, the ones responsible for the other processes essentially act intracellularly. Due to the oxidation of a variety of substrates, fungal laccases especially from basidiomycetes receive also attention for use in diverse biotechnological applications, like bioremediation. Laccases are involved in the biotransformation of many persistent environmental pollutants, like azo dyes, polycyclic aromatic hydrocarbons, endocrine disruptors or polycyclic musk fragrances.
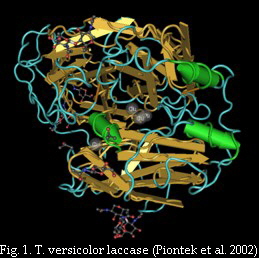
The catalytic activity of fungal laccases requires a minimum of four copper atoms per active protein (Fig. 1). Accordingly, laccases consist of four regions, which bind the copper atoms of the active centre. These copper binding regions (cbr) are strongly conserved in all laccases (Fig. 2) and were used for the design of degenerate primers to analyze and identify laccase genes of several fungal species or in environmental samples.
More informations about Laccases are available at:
The BRENDA database [link]
The KEGG database [link]
The FOLy database (Auxiliary-Activities at CAZy) [link]
The BIORENEW program [link]
During my PhD I worked within the DFG joined research program SPP 1090 "Soils as source and sink for CO2" on the topic “Diversity and expression of fungal and bacterial laccase genes in soils and cultures".
 |
|
Fig. 2. Amino acid alignment including the four well conserved copper binding regions (cbr I - cbr IV) of laccases used for the design of degenerate primers and the polyclonal antibody (based on 4 peptides). B - basidiomycetes, A - ascomycetes, Z - zygomycetes, P - plants
|
|
2. Research questions
Our main questions are of ecological origin. We wanted a molecular tool to identify fungi and assess their functional role in soils. Thus, laccase genes and their involvement in certain degradation processes were a suitable target for this kind of studies. One main aim was to characterize the genetic and functional (“expression”) diversity of fungal laccase genes in a brown forest soil in space and time. Moreover it was interesting to analyze the potential role of bacterial laccase-like genes. Consequently in future attempts it is necessary to develop proteomic approaches to investigate the role of laccases in soils.
Beside analyzing the role of laccases in the environment, I’m also interested in the genetic diversity, their expression and function in single fungal groups or taxa. Especially ascomycetes are very understudied, and need a focus in future research.
3. Methods
In order to investigate fungal laccases in pure cultures or environmental samples, standard molecular methods were used. In particular, DNA or RNA (later transcribed in cDNA) was extracted from samples and thereafter used in PCR for amplification of fungal laccase gene fragments. The amplified gene fragments, situated between the conserved copper binding regions (Fig. 3), were cloned and subsequently sequenced. Afterwards the sequences were analyzed for possible introns and aligned together. Then, phylogenetic programs were used to analyze their relationship to fungal reference species or to display the different laccase gene families. Furthermore the expression of laccases in cultures was analyzed, e.g. after induction with naturally occuring phenolic substrates. In recent attempts we try to relate the obtained genetic informations to the appearance of extracellular laccase proteins using proteomic tools like antibody incubation and mass spectrometry of tryptic fragments.
 |
|
Fig. 3. Schematic representation of a laccase gene including the conserved copperbinding regions I-IV and the corresponding primer sites for our primers.
|
|
|
Primer name
|
Primer sequence 5’-3’
|
Primer specificity
|
References
|
|
Cu1F, Primer I
|
CAY TGG CAY GGN TTY TTY CA
|
basidiomycetes, cultures & soil samples
|
D´Souza 1996; Luis 2004, 2005
|
|
Cu1AF
|
ACM WCB GTY CAY TGG CAY GG
|
ascomycetes: cultures & soil RNA (cDNA)
bacteria: cultures & soil DNA
|
Kellner 2007, 2008, 2009
|
|
Cu2R
|
G RCT GTG GTA CCA GAA NGT NCC
|
fungi and bacteria, cultures & soil samples
|
Luis 2004, 2005; Kellner 2007, 2008, 2009
|
|
Cu3R
|
TG ICC RTG IAR RTG IAN IGG RTG
|
fungi, cultures
|
Kellner 2007
|
|
Primer II
|
RTG RCT RTG RTA CCA RAA NGT
|
basidiomycetes: cultures
|
D´Souza 1996
|
|
LAC2FOR
|
GGI ACI WII TGG TAY CAY WSI CA
|
ascomycetes: cultures & soil DNA
|
Lyons 2003
|
|
LAC3REV
|
CC RTG IWK RTG IAW IGG RTG IGG
|
ascomycetes: cultures & soil DNA
|
Lyons 2003
|
|
Cu4R
|
TGC TCV AGB AKR TGG CAG TG
|
bacteria
|
Ausec 2011
|
|
Cu1Fmod1
|
ACG GTY CAY TGG CAY GG
|
fungi, qPCR on cDNA
|
Edwards 2011
|
|
Cu2Rmod1
|
G RCT GTG GTA CCA GAA IGT NC
|
fungi, qPCR on cDNA
|
Edwards 2011
|
|
|
|
|
|
|
4. Results
Within 6 years of study (Patricia Luis & me), it was possible to analyze the spatial and temporal distribution of saprotrophic and mycorrhizal basidiomycete laccase genes in a brown forest soil. Furthermore, a method to investigate their accordingly expression in soil was developed by Patricia Luis and used for the study of an annual cycle. Recent attempts were also successful to analyze the diversity of bacterial laccase genes in soil. In an cooperative attempt we successfully designed and evaluated an antibody against fungal laccases.
see detailed results of our investigations here:
Section I: Spatial distribution and expression of fungal laccase genes in a forest soil [link]
work of Patricia Luis, first phase of the SPP1090 project
Section II: Temporal laccase gene diversity and expression in a forest Cambisol [link]
work of Harald Kellner, second phase of the SPP1090 project
Section III: Proteomic detection of fungal extracellular laccases [link]
work of Harald Kellner & Nico Jehmlich, second phase of the SPP1090 project
Section IV: Spatial distribution of bacterial laccase-like MCOs [link]
work of Harald Kellner, second phase of the SPP1090 project
Further: The laccase genes diversity and expression in the Morchellaceae [link]
some “follow up” studies with laccases (as main or co-contributer):
Section V: Laccase gene expression in a sugar maple forest site, accompanied by expression of many other lignocellulolytic enzymes [link]
Section VI: Regulation of cellobiohydrolase and laccase under increased N deposition. [link] 5. Laccase gene detection & analysis in ecosystem research
A comprehensive overview of different studies is given here.
Within the last years several research projects were performed by research groups worldwide using fungal laccase genes as a tool to answer ecological questions in the field. A large focus was to answer the question how increased atmosperic N deposition alters the fungal community (here mainly basidiomycetes), i.e. laccase gene presence, abundance and composition in forest ecosystems. The group around Prof. Zak, University of Michigan used our developed primer pair (Luis et al. 2004) to analyse the abundance of laccase genes using quantitative PCR or length heterogeneity to analyse changes in different forest soils and litter after applications of different levels of NO3- (Blackwood et al., 2007; Hofmockel et al., 2007; Hassett et al., 2009; Lauber et al., 2009). The studies were performed in different sites across Michigan with different durations, one short-term study (~6 years of N application; Manistee study) and a long-term study of more than 15 years of N treatment across a wide range of sampling sites (The Michigan Gradient [link]). Recently we were able to detect a downregulation of laccase gene expression in the N amended soils (Edwards et al. 2011).
A similar research, focusing on the effects of reducing the atmospheric N input in a forest ecosystem was performed by Theuerl and coworkers (Theuerl et al. 2010).
A different study uses laccase genes to investigate the ecosystem response (i.e. fungal response) after repeated fires, which releases multiple nutrients into the soil (Artz et al. 2009; using the primers of D’Souza et al. 1996). Another research performed by the Buscot-Lab investigates the laccases genes in bulk soil and on stones (Christ et al. 2011). Recently published is Chen et al. 2012.
Parallel to studies on fungal laccases, the bacterial laccase-like multicopper oxidases gain more interest in ecological and biotechnological research. So, a tremendous amount of bacterial laccase genes were detected in peat soils and someone might ask about their ecological importance (Ausec et al. 2011).
Ausec, L., van Elsas, J.D., Mandic-Mulec I. (2011) Two- and three-domain bacterial laccase-like genes are present in drained peat soils. Soil Biology & Biochemistry 43: 975-983.
Artz, R.R.E., Reid, E., Anderson, I.C., Campbell, C.D., Cairney, J.W.G., 2009. Long term repeated prescribed burning increases eveness in the basidiomycete laccase gene pool in forest soils. FEMS Microbiology and Ecology 67: 397-410.
Blackwood, C.B., Waldrop, M.P., Zak, D.R., Sinsabaugh, R.L., 2007. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environmental Microbiology 9: 1306-1316.
Chen, X., Su, Y., He, X., Liang, Y., Wu, J., 2013. Comparative analysis of basidiomycetous laccase genes in forest soils reveals differences at the cDNA and DNA levels. Plant and Soil 366: 321-331.
Christ, S., Wubet, T., Theuerl, S., Herold, N., Buscot, F., 2011. Fungal communities in bulk soil and stone compartments of different forest and soil types as revealed by a barcoding ITS rDNA and a functional laccase encoding gene marker. Soil Biology and Biochemistry 43: 1292-1299.
Hassett, J.E., Zak, D.R., Blackwood, C.S., Pregitzer, K.S., 2009. Are basidiomycete laccase gene abundance and composition related to reduced lignolytic activity under elevated atmospheric NO3- deposition in a Northern Hardwood Forest? Microbial Ecology 57: 728-739.
Hofmockel, K.S., Zak, D.R., Blackwood, C.B., 2007. Does Atmospheric NO3- Deposition Alter the Abundance and Activity of Ligninolytic Fungi in Forest Soils? Ecosystems 10: 1278–1286.
Lauber, C.L., Sinsabaugh, R.L., Zak, D.R., 2009. Laccase gene composition and relative abundance in Oak forest soil is not affected by short-term nitrogen fertilization. Microbial Ecology 57: 50-57.
Theuerl, S., Dörr, N., Guggenberger, G., Langer, U., Kaiser, K., Lamersdorf, N., Buscot, F. 2010. Response of recalcitrant soil substances to reduced N deposition in a spruce forest soil: integrating laccase encoding genes and lignin decomposition. FEMS Microbiology Ecology 73: 166-177.
6. Laccases in Reviews
Recently several reviews were published highlighting the role of laccases in ecosystem processes and “summing up” the questions of the previous point.Kües U & Rühl M (2011) Multiple Multi-Copper Oxidase Gene Families in Basidiomycetes – What for? Current Genomics 12: 72-94.
Sinsabaugh RL (2010) Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biology & Biochemistry 42: 391-404.
Theuerl S & Buscot F (2010) Laccases: toward disentangling their diversity and functions in relation to soil organic matter cycling. Biology and Fertility of Soils 46: 215-225.
7. Further suggested readingsBaldrian P (2006) Fungal laccases: occurrence and properties. FEMS Microbiol Rev 30: 215-242.
D’Souza TM, Boominathan K & Reddy CA (1996) Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl Environ Microbiol 62: 3739-3744.
Edwards I.P., Zak D.R., Kellner H., Eisenlord S.D., Pregitzer K.S. (2011) Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLoS ONE 6(6): e20421.
Hoegger PJ, Kilaru S, James TY, Thacker JR & Kües U (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273: 2308-2326.
Kellner H., Zak D.R., Vandenbol M. (2010): Fungi unearthed: detection of expressed lignocellulolytic and chitinolytic enzymes in forest soil. PLoS ONE 5(6): e10971.
Kellner H., Luis P., Zimdars B., Kiesel B., Buscot F. (2008): Diversity of bacterial laccase-like multicopper oxidase genes in forest and grassland Cambisol soil samples. Soil Biol Biochem 40: 638-648.
Kellner H., Jehmlich N., Benndorf D., Hoffmann R., Rühl M., Hoegger P.J., Majcherczyk A., Kües U., von Bergen M., Buscot F. (2007): Detection, quantification and identification of fungal extracellular laccases using polyclonal antibody and mass spectrometry. Enzymes and Microbial Technology 41: 694-701.
Kellner H., Luis P., & Buscot F. (2007) Diversity of laccase-like multicopper oxidase (LMCO) genes in Morchellaceae: identification of genes potentially involved in extracellular activities related to plant litter decay. FEMS Microbiol Ecol 61: 153-163.
Kellner H., Luis P., Schlitt B., Buscot F. (2009): Temporal changes in diversity and expression patterns of fungal laccase genes within the organic horizon of a brown forest soil. Soil Biol Biochem 41: 1380-1389.
Luis P, Walther G, Kellner H, Martin F & Buscot F (2004) Diversity of laccase genes from basidiomycetes in a forest soil. Soil Biol Biochem 36: 1025-1036.
Luis P, Kellner H, Martin F & Buscot F (2005a) A molecular method to evaluate basidiomycete laccase gene expression in forest soils. Geoderma 128: 18-27.
Luis P, Kellner H, Zimdars B, Langer U, Martin F & Buscot F (2005b) Patchiness and spatial distribution of laccase genes of ectomycorrhizal, saprotrophic and unknown basidiomycetes in the upper horizons of a mixed forest Cambisol. Microb Ecol 50: 570-579.
Lyons JI, Newell SY, Buchan A & Moran MA (2003) Diversity of ascomycete laccase gene sequences in a Southeastern US salt marsh. Microb Ecol 45: 270-281.
Piontek K, Antorini M, Choinowski T (2002). Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-A resolution containing a full complement of coppers. J Biol Chem 277: 37663-37669.
Acknowledgement
I have to thank P. Luis, B. Schlitt, N. Jehmlich, M. Pecyna for their help as well as many more people on this topic.