|
4. Spatial diversity of bacterial laccase-like multicopper oxidases in two Cambisols
second project phase of SPP1090 in cooperation with UFZ Environmental Microbiology
Aim of this study was to develop a method to analyze bacterial laccase genes (laccase-like multicopper oxidase genes) in pure cultures as well as soil samples. As additional objective we wanted to use the same gene region (cbr I to II) as used for the fungi in our previous investigations. We screened 10 bacterial strains of different orders with our primerset Cu1Af and Cu2R and could amplify LMCO genes from 8 of them. After cloning and sequencing we found 2 different gene copies in Agromyces salentinus and Sinorhizobium morelense (Fig. 1). Database searches confirmed this multigene observation as we found different copies also in Pseudomonas putida (Fig. 1) and Sinorhizobium meliloti.
 |
Fig. 1: Amino acid alignment of bacterial LMCO genes between the conserved copper binding regions one (cbr I) and two (cbr II) compared to fungal reference sequences. Similarity shading was conducted with a 70% shading threshold in grey and 90% threshold in black. Stars mark characteristic conserved residues of LMCO genes and the triangle marks the unique cysteine residue for fungal LMCO genes. (F) accounts for sequences obtained from the forest soil samples, (G) from the grassland site, (B) from bacterial isolates and (FR) for fungal references.
However, the major question is “Can we distincly separate bacterial and fungal laccase gene sequences?”. Clearly, yes. Bacterial sequences between cbrI and cbrII are shorter (2-3 aa) and do not contain the unique cysteine residue which fungi have (there are few exceptions, mostly ascomycetes) (Fig. 1).
We examined 3 soil horizons of a forest Cambisol and one horizon of a grassland Cambisol, and found a high diversity of bacterial LMCO genes - almost without any saturation. Remarkably, some of the geneclades appear to be horizon-specific (Fig. 2). However, a gene diversity does not neccessarily mean a soil function. Thus we made some checks of pure cultures using 2,6-Dimethoxyphenol, which by literature seems to be oxidized by bacterial laccases. Seven out of ten bacterial strains were capable to oxidize 2,6-DMP. Nevertheless, expression- and proteomic studies have to be done to be sure about the involvement of this LMCO gene in the observed enzyme activity. Bacterial laccases were discussed in many different ways, to be important in physiological intracellular processes, but also biogeochemical as involved in Mn2+ oxidation. We made a quick test, and at least one of our Pseudomonas strains was capable.
Summarizing, we think there is an involvement in soil (organic matter degradation) processes, and not only in physiological processes. However, further studies are needed to show more evidence for this role.
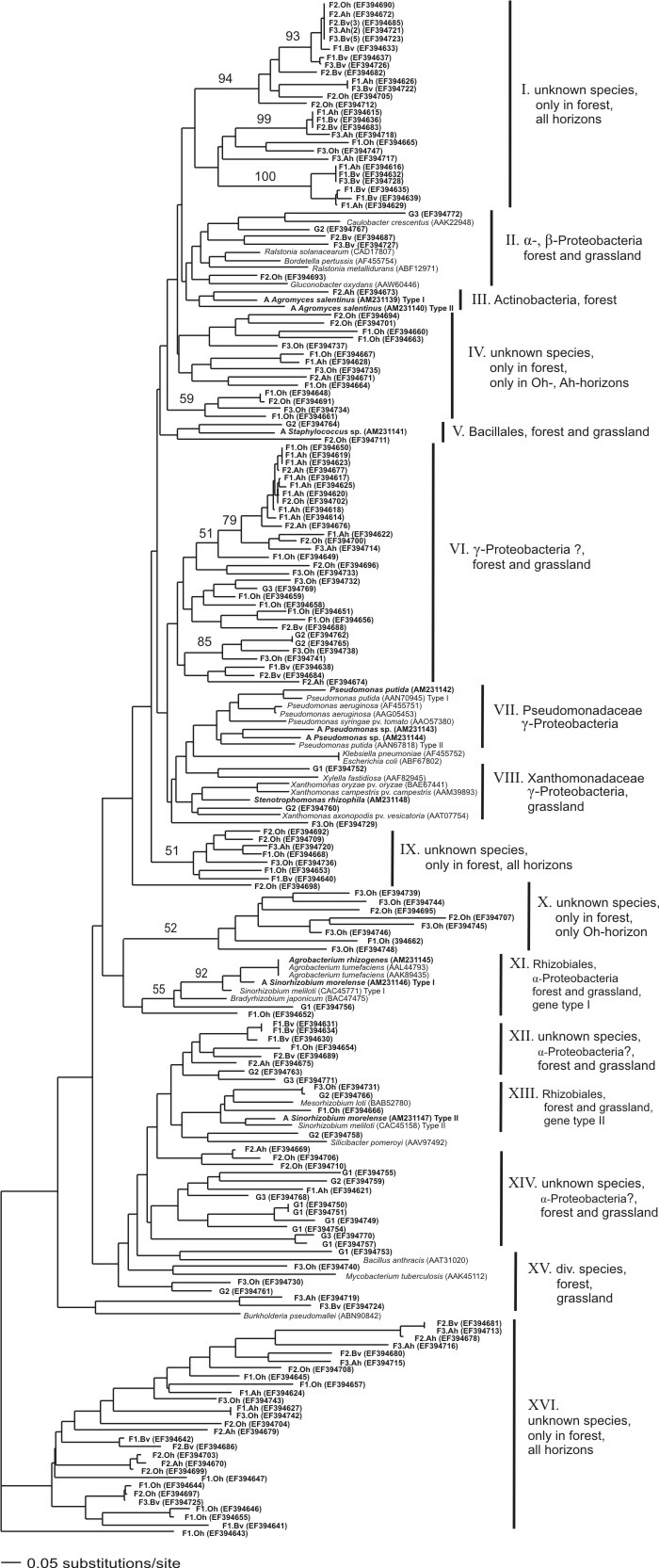 |
Fig. 2: Neighbor-joining tree representing the genetic distances of LMCO gene fragments amplified by PCR from soil samples and bacterial isolates together with reference sequences retrieved from GenBank. The NJ tree was conducted using Kimura 2-parameter distances based on an alignment of 106 positions. Values above branches indicate bootstrap support derived from 1000 replicates. (F) accounts for sequences obtained from the forest soil samples, (G) from the grassland site and (A) from bacterial isolates. The number of clones sequenced is indicated within brackets if >1.
The alignment is downloadable from TreeBASE (study no. 2008) or here [link]
Kellner H., Luis P., Zimdars B., Kiesel B., Buscot F. (2008): Diversity of bacterial laccase-like multicopper oxidase genes in forest and grassland Cambisol soil samples. Soil Biology & Biochemistry 40: 638-648.
doi:10.1016/j.soilbio.2007.09.013
|