|
1. Molecular phylogeny of the Morchellaceae
In one study the phylogenetic relationship of different morel species as well as near relatives like Verpa species and Disciotis venosa was analysed. Analyses using ITS sequences displayed a monophyletism of the family Morchellaceae as well as the genus Morchella (Fig. 1). Using ITS sequences was difficult as single species or groups showed remarkable differences in the sequence length. For example, the black morel M. conica had an ITS sequence of 737bp whereas the yellow morel M. crassipes had 1226bp. This made an alignment difficult. Nevertheless the alignment includes altogether 321 positions; the complete 5.8S rRNA gene and parts (domains) upstream.
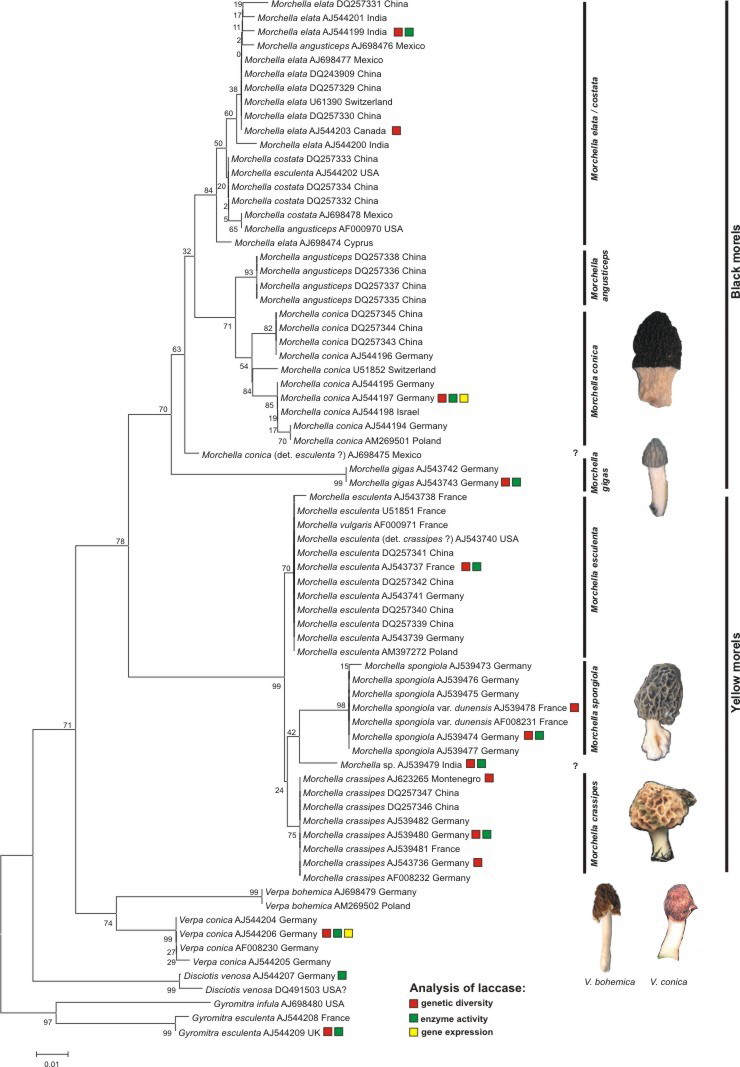 |
Fig. 1. NJ-tree using K2P-distances. Supportvalues were received using 2000 bootstrap replicates. The alignment of the ITS-region included 321 positions. Species analysed for their laccase-like multicopper oxidases were marked by a color code. Fruitbodies displayed here are from german populations.
Thus, using ITS sequences for recognition of species clearly separated four yellow morel species (M. crassipes, M. esculenta, M. spongiola and one undescribed morel from India), then the half-free morel M. gigas which clusted within the black morels and finally a third group including the remaining black morels (M. elata/conica/costata - clade). The latter group seems to be very diverse and has to be investigated more detailed in future. Nevertheless a separation into two main clades (M. conica and M. elata) is possible using the full length ITS sequences (~740bp) (data not shown).
In a second study an analysis of partial LSU (28S rRNA gene) sequences was conducted to resolve the monophyletism of the Morchellaceae. This was not supported (no bootstrap support) using ITS sequences, due to their length variability. A bayesian analysis as well as a maximum parsimony approach with bootstrapping showed strong support of the Morchellaceae, the genus Morchella and the separation into black morels (sectio Distantes) and yellow morels (sectio Adnatae). Again, M. gigas (half-free morel) formerly treated as species of the genus Mitrophora, distincly clustered in the black morels. They therefore could also assumed as black morel (sectio Distantes), but in a extreme manner.
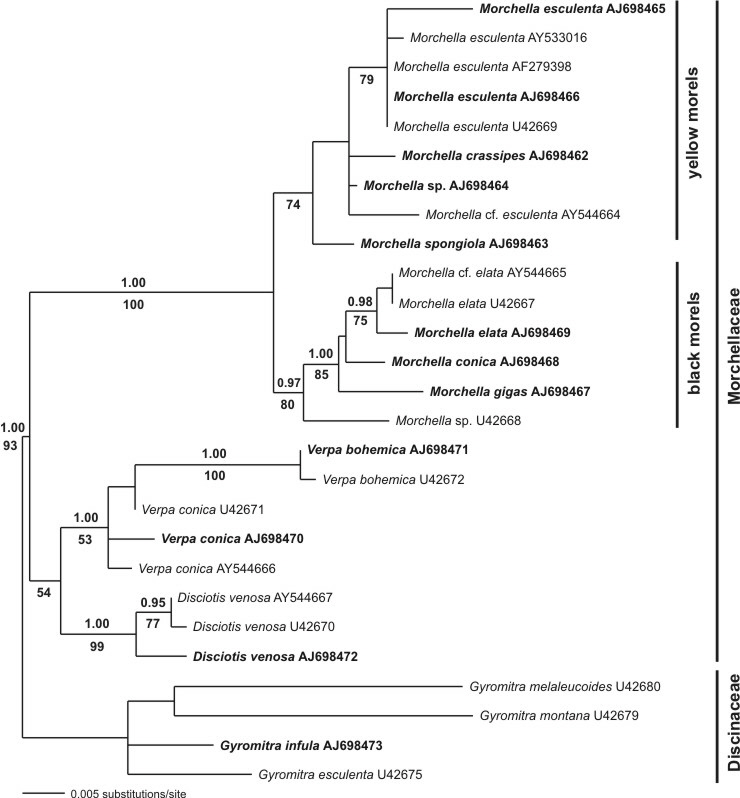 |
Fig. 2. Consensus tree using LSU sequences and a bayesian approach (a-posteriori probabilities above branches). Furthermore a Maximum Parsimony approach using 2000 bootstrap replicates was conducted (values below branches indicate support).
All analyses are available. The used sequences are available in GenBank [Link]. Furthermore the used alignments are available at TreeBASE (study no. 1748) [Link].
But the used alignments can also be downloaded here:
|
Gene / Region
|
Species
|
ntax
|
nchar
|
click to download
|
|
ITS
|
all Morchellaceae
|
74
|
1311
|
[.nex] [.fas]
|
|
ITS
|
only black morels
|
33
|
752
|
[.nex] [.fas]
|
|
LSU (28S)
|
Morchellaceae
|
22
|
1131
|
[.nex] [.fas]
|
|
All alignments include full sequences. Excluded regions within subsequent analysis are indicated! I used PAUP and BioEdit for generation and analyses.
|